Into The Ocean
Environmental DNA: what is it and how can we use it for coastal plant conservation?
Recently, I published a scientific paper from my PhD research titled "A Multi-Gene Region Targeted Capture Approach to Detect Plant DNA in Environmental Samples: A Case Study from Coastal Environments". If you don't think that sounds
like a nice pre-bedtime read that's okay. Scientific articles are generally targeted to audiences in the specific field of
which they are published. In this case, I wrote with the intention that scientists working in the field of environmental
DNA would read this article. However, as I shared this article with friends and family, I realised it was not easily
digestible to people not in the field. That is where this blog comes in, I am going to write about this research paper,
provide background and share the key findings with hopefully less technical words.
Firstly, let's start with the basics, what even is environmental DNA? Every organism in the world leaves behind a piece
of themselves wherever they go, we shed hair, skin cells and by extension, DNA, everywhere. If you have seen the movie
GATACA starring Ethan Hawke and Jude Law, then you would be familiar with how easily DNA is shed given the rigorous
self-cleaning routine Vincent went through to cover up the evidence that he was genetically inferior. Fortunately for us,
this accessible DNA can be used to assess plant and animal presence in environmental samples such as soil, water and even
excrements. This is termed environmental DNA and is the DNA left behind by organisms within environmental samples.
So, we know what environmental DNA is, and from the paper heading you would see I am specifically looking at plant DNA in
environmental samples but what is a targeted capture approach? what does multi-gene region mean? and what does this have to
do with coastal environments!?!? Before we panic let's take it slow and start by discussing how we actually analyse
environmental DNA. The first step in analysing environmental DNA is to get it out of the environmental sample or as we
say, "extract the DNA". Fortunately, some very clever people worked out how to do this a while ago and now I can buy an
extraction kit and follow the instructions. It is just like buying one of those cheat cake boxes with all the ingredients
and instructions to follow and voila you have a cake; except this time, it is DNA.
Alright, now the next part is tricky. I don't need to tell you that plants are different to humans, and they have something
that we do not, they have a chloroplast. This is what is responsible for the neat photosynthesis trick that they like to do
which keeps us alive. To photosynthesise, plants undergo a chemical process and in order to do this they need the right
proteins and in order to make the right proteins, they need the right set of instructions which is packaged up into what we
call, genes. Each gene within chloroplasts encode for a different protein and between different plants the genetic code in
certain genes is slightly different. This is very helpful to the identification of plants as this is basically a unique code
that identifies the plant ‑ almost like a barcode…actually, exactly like a barcode, as this is the scientific term for it.
The unique barcodes within plants have already been identified by some super smart cookies some time ago. To compare
barcodes (gene regions) between different plants, this region needs to be found within the DNA and multiple copies of this
exact region are required so that the region is easily identified. This all occurs through a process called PCR or Polymerase
Chain Reaction. This is the super cool method of amplifying regions of DNA and is likely one of the most important molecular
processes that we have. PCR works by the design of PCR primers, which are small regions of DNA that are designed to bind to
DNA just outside of the region of interest i.e., on either side of the barcode region. The addition of DNA polymerase (an
enzyme responsible for replicating DNA) enables the so-called target region or barcode to be replicated millions of times,
thus creating millions of copies of this region to study.
Once PCR is complete, the amplified region then gets sequenced... DNA sequencing determines the coding amino acids in DNA ‑
AGCT (Adenine, Guanine, Cytosine and Thymine) and once sequencing is completed, a file is created specifying the specific
AGCT configuration of the target gene region or barcode. This is then what gets compared between plants with certain plant
groups possessing a unique combination of characters. The chosen DNA barcode can either be conserved or variable between
plant species. Conserved regions do not change much over time and are thus similar or the same between many plant groups.
Variable regions are those which are constantly evolving over time and are therefore much more unique between plants ‑ it
is these regions that are desirable for identifying plants in environmental samples.
Universal barcode regions are genes that are different between many plant groups, and these are highly desirable for
environmental DNA work. This is called metabarcoding as it uses the amplification of one gene region to identify many
species. The downsides of metabarcoding are that only one gene region can be analysed at a time and while this is chosen
to be variable between many different plant groups, often this will only identify families of plants, not the species ‑
which is more useful. Alternative gene regions that identify plants to species level are much longer and thus more difficult
to amplify from environmental DNA. Why might this be? Because once DNA is released into the environment certain enzymes
and microbes break down this DNA into smaller pieces. The smaller the DNA the less likely there will be intact sites for
primers to bind to and therefore this region will not be amplified for all plants within the sample. This leaves us with
a conundrum, how can we improve environmental DNA analysis to recover all of the plants present within an environmental
sample with enough information to identify which specific species are present.
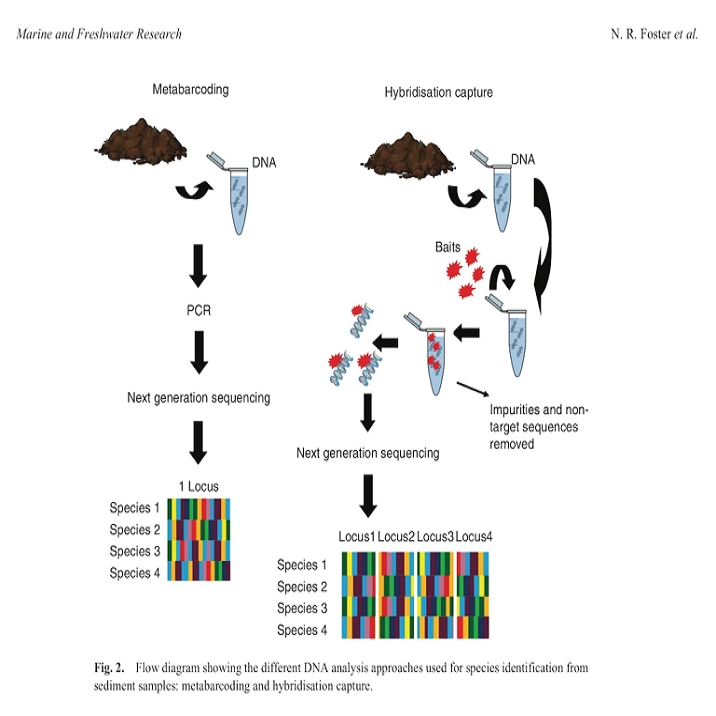
Well, we finally made it! This is where my research comes in (from here on out I will be referring to my research as "we"
or "our" given this was a team effort). We developed an approach that uses targeted capture rather than primer binding to
amplify DNA from environmental samples. What does this mean? New technology has been developed where we can now design what
are called "baits" and this are small pieces of RNA than bind to DNA just like primers do except they don't rely on intact
primer binding sites to amplify DNA and there are many thousands of target regions instead of just the one. This means we
can recover multiple gene regions and increase the likelihood of identifying all plants present in the environmental
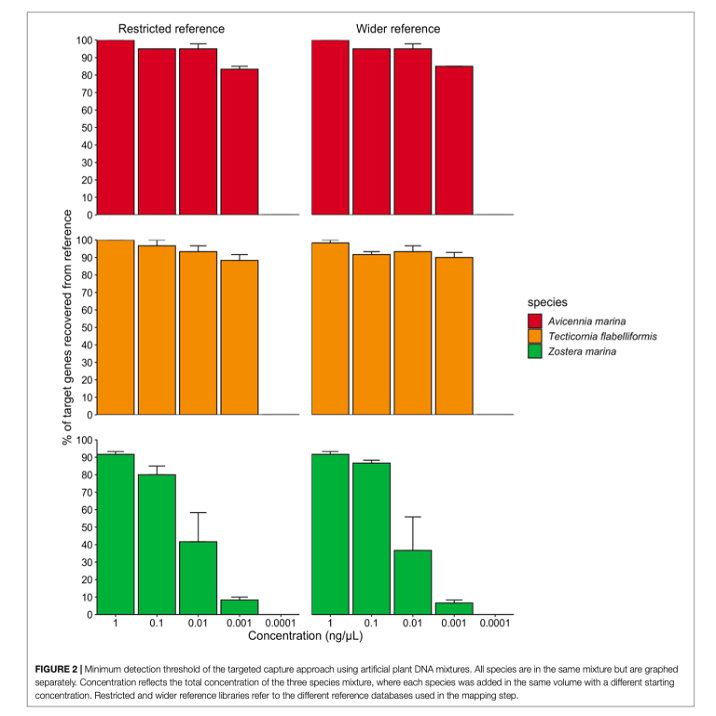
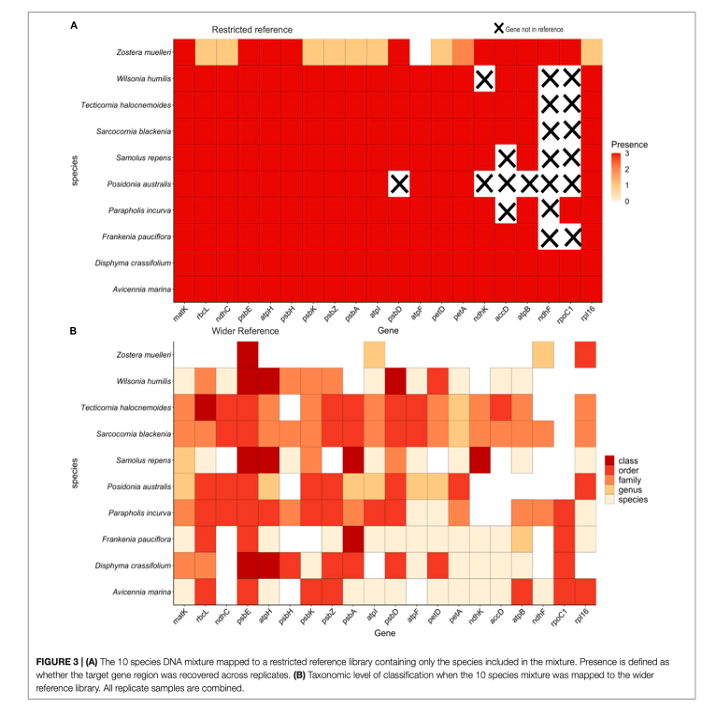
sample and decipher these to species identification. The moral of this paper is... it worked! We managed to use this
approach on mock DNA mixtures and recover all species we knowingly put into the sample across 90% of the genes that we
targeted and down to low levels of DNA concentration. This means we improved both the detection ability of this method
and the ability to identify plants with higher accuracy (multiple genes increases the amount of genetic information to
separate species). We also tested this approach on actual environmental samples and could recover plant species known to
inhabit the area.
If you have read this far, great! We are almost done. You may be wondering well why should I care about this? Why is
environmental DNA important? Why is research being done to improve the ability to detect plant DNA in environmental
samples? Well, environmental DNA can be extremely useful to environmental research. We can collect sediment samples from
deep underground that were buried many years ago and examine what sort of plants were growing in an area hundreds,
thousands and even millennia ago. This is useful to understand species extinctions, introductions and the impacts humans
have had on our planet. By looking into past plant communities, we can better predict what is going to happen in the future
as we can observe the survivors of previous environmental disasters and the mechanisms for a system to recover from a
disturbance event. Environmental DNA offers a unique glimpse into the history of planet earth and the learning opportunities
from this are endless.