Into The Ocean
What is a plant barcode?

Picture this, you head to the supermarket to purchase food for a week, you collect the items you need, and if you are
like me this includes several additional "treats", and you then wander over to the checkout. Each item is scanned by a
barcode reader, the computer reads the barcode and records the price, item after item is scanned and then suddenly, the
production line halts, the barcode cannot be read, you're in trouble. The computer cannot understand what the item is,
it is missing the unique code that has been assigned to all the other items. If this is familiar to you then you already
have a basic understanding of DNA barcoding but replace barcodes with sequences of DNA and items with plants. DNA barcoding
is an extremely important field of research, enabling the identification of plants within our environment. After DNA is
extracted from plants uniquely identified gene regions or barcodes are amplified using Polymerase Chain Reaction ‑ this
is the process by which DNA is amplified. The resulting amplified DNA is then sent to be sequenced i.e., the DNA sequence
is read and a whole bunch of A's, G's, C's and T's are recorded. The combination and order of these four characters
dictates the unique barcode for each plant species on earth.
Generating large databases of these unique DNA sequences is a challenge and currently databases are severely depauperate,
meaning there are lots of plants with missing barcodes. This is a serious problem in the field of environmental DNA, which
is the recovery of DNA from environmental samples such as water and soil (mentioned previously). If DNA is recovered from
an environmental sample to say, try to detect an invasive species, monitor diet or document diversity, then this sequence
needs to be matched to a database, otherwise it is just a series of characters with not much use. The recent study I worked
on looked at a method to generate these barcodes for a large number of plant species across multiple barcodes. This is where
it gets tricky, as plants can have multiple barcodes, not just one. This is because not all barcodes are as unique to the
species, genus or family as others, and so the ability to uniquely identify a plant may depend on one or a combination of
plant barcodes. This means not only do we need to increase the number plants which have a barcode sequence generated, but
also increase the number of these barcodes for each species to ensure unique identification. Not an easy task that's for
sure! but recent advances in both sequencing and the advent of targeted capture (explained here) is making this more
achievable.
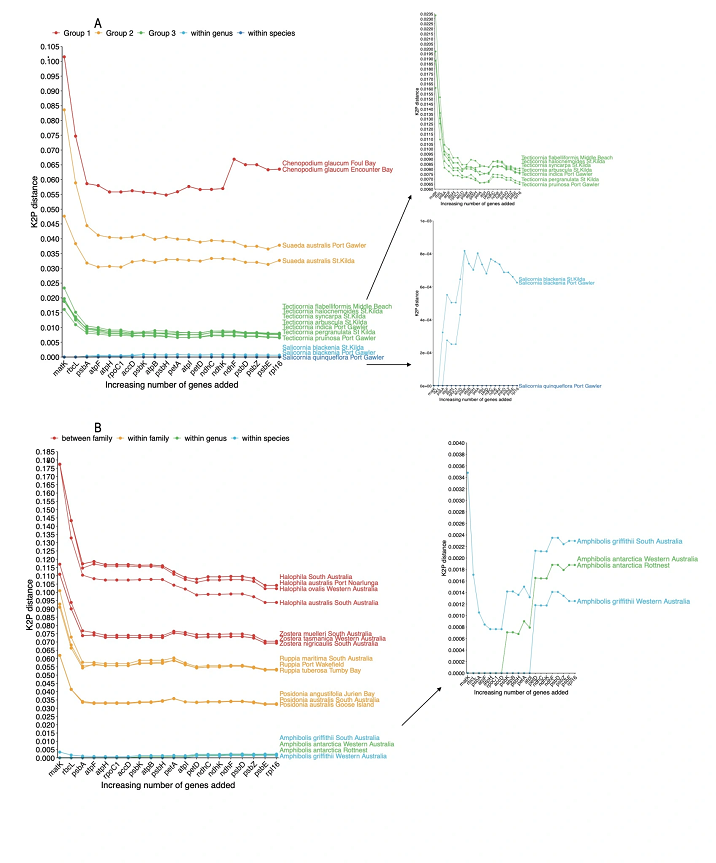
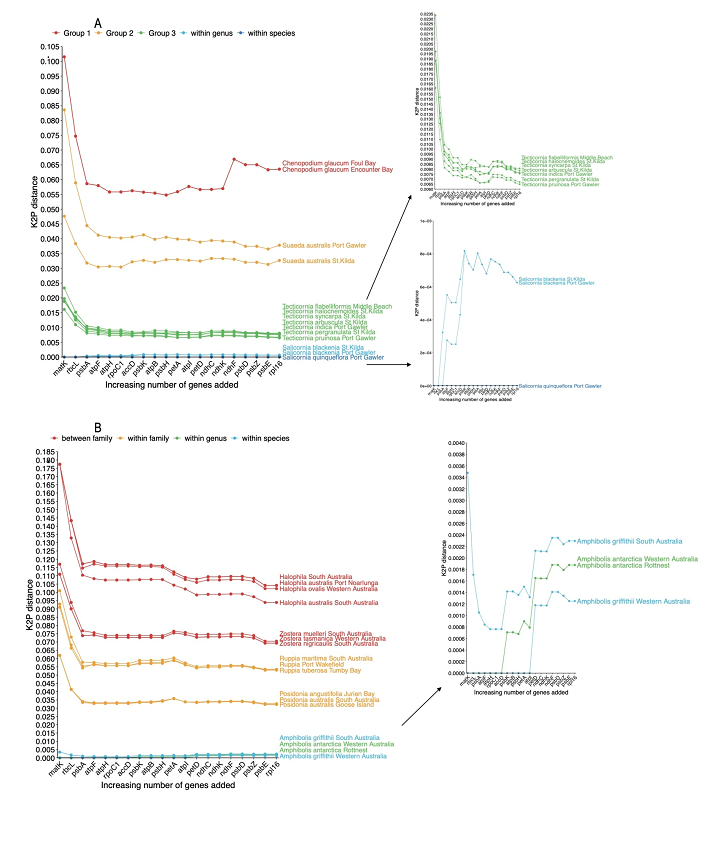
In my recent research published in Ecology and Evolution, I applied targeted capture to generate 20 barcodes across 93
plant species, all of which were coastal temperate plants, given my field of interest. I had a 92% success rate for recovering
all targeted barcodes across the 93 samples. I also investigated which of the barcodes may be better to use to identify
species, in other words, which barcode was unique to most of the species in the database. I found this varied across
different plant families and the combination of multiple barcodes was the best way to uniquely identify closely related
species. This approach to barcode generation for plants means multiple barcodes can be developed for the same cost and
time to generate one. So in the race to generate barcodes for all flora on earth this research leads the charge and is a
great step towards barcoding all plants, an initiative that is currently being undertaken for all of Australian flora within
the genomics for Australian plants project and the Tree of Life Initiative.